27 June 2016
Making trees, or; The triumph of molecules
One more quick observation from the Evolution meeting in Austin last week.
Every tree of relationships between species I saw was based on molecular data.
This interested me, because there was a point where there was controversy about whether DNA data could be used to make phylogenies. I seem to remember articles that argued that relationships based on morphological data would generally be superior to those from DNA.
Now, I wonder if the tables have turned so much that if you put up tree based on morphological data instead of molecular data if people would look at it funny. (There would be a few exceptions, obviously, like fossil data.)
Photo by Extremely Tropical on Flickr; used under Creative Commons license.
22 June 2016
Evolution 2016, Day 5
Perhaps the highlight of my last day of the Evolution meeting was learning from Paul Durst that you can get fleas off a (dead) mouse by putting the body in a paint can shaker. The next speaker, Matthew Forister, started his talk by saying, “Dead mice in a paint shaker. Tough act to follow.”
Forister also won the “Overly honest methods” award for saying at one point, “Does this have anything to do with the real world? I don’t know.”
Some final impressions of this meeting.
 I was surprised by how humourless the talks were. Dead mice in a
paint shaker notwithstanding,very few people tried to make a joke. I
only saw one Star Wars quote in a talk, even though a social game for students, Evolution bingo, encouraged people to put Star Wars quotes in their talks. I didn’t see any clown noses in the poster sessions, either.
I was surprised by how humourless the talks were. Dead mice in a
paint shaker notwithstanding,very few people tried to make a joke. I
only saw one Star Wars quote in a talk, even though a social game for students, Evolution bingo, encouraged people to put Star Wars quotes in their talks. I didn’t see any clown noses in the poster sessions, either.The food and drink was very good. I’ve been at some small meetings that had snacks this good, but not a conference that was this big.
This was probably the smoothest running set of presentations I have ever seen. While I made fun of the chimes, they worked better than conference moderators. People started and stopped on time.
The precision timing of the talks mattered little, however, when you have one talk in room 10-C and the next one in Ballroom C. The distance between them was long enough that it took probably 3 minutes to get from one end of the meeting rooms to the other, so you were invariably missing the starts of talks.
On a related note, sixteen concurrent sessions is too many.
When you see a lot of cool talks using the same sort of organisms, you feel like the stupidest person in the world for not working on them. “Why am I not studying guppies? What is wrong with me?!”
External links
Evolution 2016 bingo card
21 June 2016
Evolution 2016, Day 4
Yes, I saw some cool science, yesterday, including some cool and contradictory results on how predators shape brain evolution (big brains are favoured in high predator environments in guppies but not killifish?) and jump right to the big news.
The Society for the Study of Evolution's flagship journal, Evolution, will be moving to an online only journal, with all papers becoming open access two years after publication. Decades of papers, including many classics, will be free to read in early 2017.
The Society is also launching a new online, open access, "high impact" journal in early 2017, Evolution Letters.
Let me be among the first to congratulate the Society for the Study of Evolution for moving their publications toward a superior and more modern way of scientific communication.
And I think I am among the first to pat the Society on the back, because, judging from the reaction in my Twitter feed, these announcements are widely regarded as bad moves.
People are mad that Evolution won't be immediate open access, that the two year embargo is too long for NIH funded researchers, and that the journal is still being published by Wiley, one of the biggest for profit publishers.
People (including, it must be said, myself) worry that Evolution Letters might as well be titled Evolution Rejects. The perception is that the journal will be a dumping ground for those papers that are not considered novel enough for the flagship journal.
I've been critical before about the creation of new journal that serve no editorial purpose. I worry Evolution Letters will be one of those. It's not being created to define an emerging field of research, but as part of a business plan. But I have no doubt that it will have an audience. Scientific manuscripts expand to fill the available journals.
I worry that the "54 40 or fight" attitude to open access might be a little counterproductive. While It's important shift that Overton window, it might be that criticizing good but imperfect progress might discourage people from trying to make any progress at all.
20 June 2016
Evolution 2016, Day 3
Obviously, the biggest, most important day at any conference is the one where you present your own research. 😉 First, thank you to all who came and talked to my student Claudette and me about sand crab eyes! The poster is up at figshare, along with some others from the meeting; search for "evol2016".
Observations from the second full day of the conference:
Some rooms were consistently standing room only (afternoon sexual selection session), while other, much bigger rooms were nowhere near capacity.
Lightning talks. There were a lot of these Ignite! style, short presentations. While I love this format, I think that the vast number shows of lightning talks, combined with the funkiness in room size, shows that Evolution is at a transition point in size. I think it's getting too big for talks to be the norm, and in the next few years it will start to transition more to a more poster oriented meeting.
A few talks I saw - just a random sampler, not in any order:
Aaron Owen showed the first example of rapid evolution (over a couple of centuries) in a mammal, an Indian mongoose. This was a finding that was worth having to look at mongoose butt for.
Trevor Fristoe showed that big brains allowed birds to invade variable, harsh climates, rather than harsh climates selecting for big brains.
Speaking of brains, Alberto Corral-Lopez showed that female guppies with big brains were much better at selecting sexy males with lots of colors than their small brained brethren.
Frances Hausers and Sarah Dungan both presented nice work on opsins. Dungan's talk introduced me to "Concept I have to look up and read more about when I get back": intramolecular epistasis. In my notes next to the phrase: "This seems kind of important." Not going to try to unpack it in a morning after blog post. (Frances was a fan of the Better Posters blog, too!)
Online people I got to meet in person included Scientist Sees Squirrel blogger Stephen Heard, and chat at the pub with gif master Dr. Rubidium.
19 June 2016
Evolution 2016, Day 2
The conference began in earnest Saturday, and some of the notable things were about the conference organization rather than the science.
Bells and chimes. To keep speakers on time, the conference is not relying on unreliable moderators. Each talk begins with a "Setting sail" ship bell. Near the end, there's a doorbell sound that makes me want to say, "Avon calling!" In every talk. Sometimes they chimes are are subtle, and sometimes they'll make you bolt out of your seat and yell, "YES MA'AM I'M AWAKE THE ANSWER IS TWENTY!!"
The conference also has the slickest speaker interface for slides I have ever seen. Instead of a series of strewn out files on the desktop, there is a single clean custom interface with talks and speakers clearly labeled.
The talks are kind of strewn out all over the convention center, and can make it hard to hop between talks on time. Ballroom C is a long way from room 10A! And the difference in room size is puzzling. Some rooms seems to seat 50, while the biggest ballrooms seem to seat 500 or more.
LIkewise, there are long keynote style talks and Ignite style talks interspersed in some rooms, with no clear reason for the variation. It does break up the pacing, but makes it harder to plan how to transition between rooms.
I'll have more to say about the poster session in the Better Posters blog.
Some scientific highlights included a talk by Naomi Pierce on ant symbiosis, mainly with caterpillars, but more recent work with bacteria. Yes, people, the microbiome is one of the hot and inescapable topics at this conference. Very fun stuff.
Scott Solomon gave a talk plugging an upcoming book called Future Humans. I think this book will fill an important niche, because I see so many people outside of biology who start biology questions with, "Because humans aren't evolving any more..." NOPE. Solomon's book will be the one stop source to point to people to show what we know about current human evolution.
I saw a lot of talks on sensory systems (I am allegedly a neurobiologist, after all). There were interesting talks on the variation of eyes in butterflies. And an intriguing talk that showed that eye size alone seemed to provide a fitness advantage in water fleas.
I was super pleased to meet some people I knew from social media for the first time, like Jeremy Yoder and Lenny Teytleman.
And there is the opening chime for Day 3! Time to stop blogging and start note taking!
18 June 2016
Evolution 2016, Day 1
I've wanted to go to the Evolution meeting for years. I was so delighted when it practically landed in my backyard this year in Austin.
Of course, because Texas is big, it probably too me longer to travel by car to the meeting from within the state than many others took by flying in. That last mile on I-35 is always a killer (15 minutes to go about a mile).
The opening evening started with a social, where two former UTPA students came up and found me - one who'd been in one of our undergraduate training programs and is finishing up a master's at Michigan, and one who'd done a master's and is now doing a Ph.D. here in Austin. It's so good to see people who are continuing!
Carl Zimmer gave the opening talk, after receiving the Stephen Jay Gould award for writing about evolution. Carl talked about how successful Gould was as a writer, and of his one encounter with him. Carl was working as a copy editor for Discover magazine, and got asked to edit one of Gould's essays. Now, Carl didn't say this, but Gould was infamously protective of his prose. Gould was many things, and even his close friends admitted he could be difficult. Carl said, "I think I got him to change one comma."
Carl's talk mostly focused on the discoveries around human evolution, which have so emphatically shown a point that Gould made again and again: evolution is not a straight line march of progress, but a "luxuriant bush." But Carl noted, rightly I think, that even Gould would not have predicted the wild interbreeding that ancient DNA evidence is showing us. "Call it a bramble, call it an orgy...".
I saw a few tweets suggesting that Carl's talk was too basic for a scientific conference. He did introduce DNA, in case someone had forgotten it. What I think some people forgot was two things.
First, there were a lot of students in the room. A lot of this information would be new to them, or maybe something that a quick reminder of basic s would help them follow.
Second, and I may be mistaken here, I think this talk was intended as a free talk, open to the general public, not just for conference attendees. And there I think it would have been completely appropriate to mention some of the basics that Carl did.
Today's sessions are starting in about six minutes. I hope to have a day 2 report tomorrow!
10 June 2016
Neurons older than dinosaurs, and homologous cells
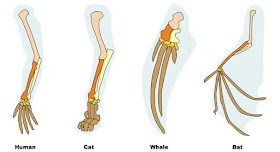
If you look at your arm, you have a single long bone connected to your shoulder, followed by two more long bones, a bunch of little bones that make up your wrist, and a set of fingers.
It’s the same in all the vertebrates. Why that particular arrangement of bones? They’re all inherited from a common ancestor. In technical terms, those bones are homologous.
I was thinking about homology after writing this answer on Quora to the question, “What is the most fascinating thing you know about in the field of neuroscience?” Here’s my reply, inspired by my 2008 paper::
After writing this answer, Namnezia pointed out that ion channels are even older. Ion channels are molecules, and it struck me:
We talk about homologous organs, like the arm bones I mentioned above, all the time in biology.
We talk about homologous molecules, mainly sequences of DNA and the proteins they make, all the time.
But the escape neuron answer I gave above may be a little unusual because I am talking about single, identifiable cells as homologous. It struck me as interesting that we routinely talk about homology in levels of organization above cells, and below cells. But outside of identified neurons in neurobiology (and invertebrate neurobiology at that), people rarely talk about homology at the level of single cells.
The only other examples of cells that people called homologous were few. You can talk about homologous cells in C. elegans and other nematode worms, because those small animals have predetermined cell fates in development, and we can map every single cell.
Is the cellular or tissue level of organization different from molecules or organs in terms of our ability to identify distinct homologs? Or have we just not looked closely enough?
Reference
Faulkes Z. 2008. Turning loss into opportunity: The key deletion of an escape circuit in decapod crustaceans. Brain, Behavior and Evolution 72(4): 251-261. http://dx.doi.org/10.1159
/000171488
Picture from Anaspides, a Living Fossil
It’s the same in all the vertebrates. Why that particular arrangement of bones? They’re all inherited from a common ancestor. In technical terms, those bones are homologous.
I was thinking about homology after writing this answer on Quora to the question, “What is the most fascinating thing you know about in the field of neuroscience?” Here’s my reply, inspired by my 2008 paper::
You can identify neurons that are older than dinosaurs.
Not the actual live neurons in an individual, of course, but types of neuron has been inherited for hundreds of millions of years.
Shrimp, crayfish, clawed lobster all have giant neurons that they use to perform escape movements, called lateral and medial giant neurons.
Now, this little guy is Anaspides:
It has lateral and medial giant neurons, too, and it uses them to perform escape movements.
While some call this species a shrimp, it isn’t a shrimp like most people are familiar with. The shrimps and prawns most people know are decapod crustaceans, and Anaspides isn’t a decapod. (This is why you don’t use common names for scientific purposes.) The oldest decapod fossil is about 350 million years old, so these escape related giant neurons must be older than that.
I think it’s awesome that you can identify neurons in species that are separated by at least 350 million years of evolution. It makes me appreciate deep time.
After writing this answer, Namnezia pointed out that ion channels are even older. Ion channels are molecules, and it struck me:
We talk about homologous organs, like the arm bones I mentioned above, all the time in biology.
We talk about homologous molecules, mainly sequences of DNA and the proteins they make, all the time.
But the escape neuron answer I gave above may be a little unusual because I am talking about single, identifiable cells as homologous. It struck me as interesting that we routinely talk about homology in levels of organization above cells, and below cells. But outside of identified neurons in neurobiology (and invertebrate neurobiology at that), people rarely talk about homology at the level of single cells.
The only other examples of cells that people called homologous were few. You can talk about homologous cells in C. elegans and other nematode worms, because those small animals have predetermined cell fates in development, and we can map every single cell.
Is the cellular or tissue level of organization different from molecules or organs in terms of our ability to identify distinct homologs? Or have we just not looked closely enough?
Reference
Faulkes Z. 2008. Turning loss into opportunity: The key deletion of an escape circuit in decapod crustaceans. Brain, Behavior and Evolution 72(4): 251-261. http://dx.doi.org/10.1159
/000171488
Picture from Anaspides, a Living Fossil
09 June 2016
Hello Atlantic! Here are my answers to your questions about funding
 The Atlantic recently ran an article about science funding that asked for scientists to write in and answer three questions. So, I did. These were my answers.
The Atlantic recently ran an article about science funding that asked for scientists to write in and answer three questions. So, I did. These were my answers.How did those years of flat (National Institutes of Health, NIH) funding affect you or your colleagues, if at all?
The difficulty in attracting funding affected our students. When I took students to conferences who were considering looking for doctoral positions, they were disheartened by how much of the talk was about whether a lab could get funding.
Flat NIH didn’t affect me or my colleagues much, for two reasons.
- My university is an emerging research institution, so there is not a huge amount of federal funding in general. Faculty here haven’t reached the point of being competitive for the stand alone R01 research grants that are the bread and butter of many biomedical research labs.
- My department is not a biomedical department. The National Science Foundation and other organizations are generally better funding fits than the NIH for us. (And it is a bit annoying when reports treat the NIH as if is was the only funding agency for all of biology. There are huge swathes of biology that NIH doesn’t touch.)
Have you even noticed the 2016 increase?
Nope.
(Other researchers often get annoyed at me when I say declining funding rates haven't affected me personally. They badly want to show solidarity, and impress on people that the funding shortfalls are hurting science – which they are, and I agree with. But it doesn’t affect all of us equally.)
And what would more funding mean to you?
My institution is determined to add many new doctoral programs, including biology. I’good thing that would creating new opportunities for underserved minority students in my region, instead of this:
ve had many discussions with my colleagues about whether this is a good idea, given the steady decline in funding success rates. More funding might convince me that a new doctoral program could be a
Additional: Moments after I posted this complaining how NIH is so often presented as the only game in town for biology funding, what do I see but a tweet from the Society for Neuroscience presenting NIH as the only game in town for biology funding. Sheesh.
Contact your representatives today and ask them to make the case for a strong research funding level for NIH.
If you’re going to all the trouble of contacting your federal representative to support neuroscience, why not mention other agencies that fund that discipline? Like the National Science Foundation?
Related posts
Happy sequestration
External links
NIH Funding: It’s Personal
08 June 2016
The cages we scientists make for ourselves
“We need to change incentives!”
Ah, how many times I have heard some variation of that phrase in describing scientific publishing.
With the creation of UTRGV, my department was forced to create new evaluation documents for annual review, for merit and tenure, and so on. Creating policy documents sounds dull, but I was quite excited by this. You don’t get many opportunities the scrape away all the junk that accumulated over the past few decades that nobody could be bothered to change. This is not an opportunity that comes along every day.
I argued to change our department’s incentives structure. I had a few things I wanted to accomplish.
- I wanted us to reward open access publication and data sharing.
- I wanted to broaden the range of things that could be considered scholarly products to include more than journal articles.
- I wanted our evaluation document to reflect that the current world of scientific publishing is largely online.
My arguments did not convince my colleagues. Mostly.
People voted in favour of rewarding people for editing a book (which was previously missing from our list), or getting a patent. Progress!
People did not vote in favour of reward sharing datasets (e.g., on Figshare) or computer code (e.g., on Github), although those votes were close. Promising.
The discussion over rewarding publication was revealing.
Previously, we had given multipliers for whether a paper was published in a regional, national, or international journal. I proposed that instead, we give more weight for an open access journal article, and less weight for an article that appeared in a print only journal (e.g., not available online).
There were two arguments against rewarding open access papers.
The first was “But it costs money.” I pointed out that many open access journals charge nothing, or have fee waivers. I was also not sure why “I have to pay” was seen as a problem, since one of the legacy departments has long rewarded people for each scientific society they belong to, and that’s an out of pocket expense to get a reward, too.
The second objection was prestige. I provided links and papers to support the arguments of the benefits of open access, the pitfalls of Impact Factor, and that reprint requests don’t cut it compared to genuine open access. But they were not swayed.
Ultimately, it felt like asking my colleagues to image a world where a PLOS ONE paper was worth more in an evaluation than a Nature paper was like asking them to picture a reddish shade of green. They just couldn’t imagine it.
The department voted against the new multipliers.
So the next time you hear, “We just have to change incentives for scientists,” remember that these existing incentives are often ones that many scientists actually want. They are in a cage of their own making and could leave at any time, but won’t.
Photo by Amber Case on Flickr; used under a Creative Commons license.